

Scientific intelligence platform for AI-powered data management and workflow automation


Scientific intelligence platform for AI-powered data management and workflow automation

Objective: The objective of the study was to evaluate the utility of a Petrifilm-based on-farm culture system when used to make selective antimicrobial treatment decisions on low somatic cell count cows (<200,000 cells/mL) at drying off. A total of 729 cows from 16 commercial dairy herds with a low bulk tank somatic cell count (<250,000 cells/mL) were randomly assigned to receive either blanket dry cow therapy (DCT) or Petrifilm-based selective DCT.
Year: 2014
Source: Journal of Dairy Science
Link: http://www.sciencedirect.com/science/article/pii/S0022030213007352
Clinical Area: Veterinary
| Sample Size Section in Paper/Protocol: |
|
“Sample size calculations were based on a published prevalence of IMI at calving |
Summary of Necessary Parameter Estimates for Sample Size Calculation:
| Parameter | Value |
| Significance Level (2-Sided) | 0.05 |
| Expected Prevalence Difference | -0.05 |
| Control Group Prevalence | 0.2 |
| ICC | 0.2 |
| Sample Size per Cluster/Cow | 4 |
| Cluster/Cows per Group | 454 |
Step 1:
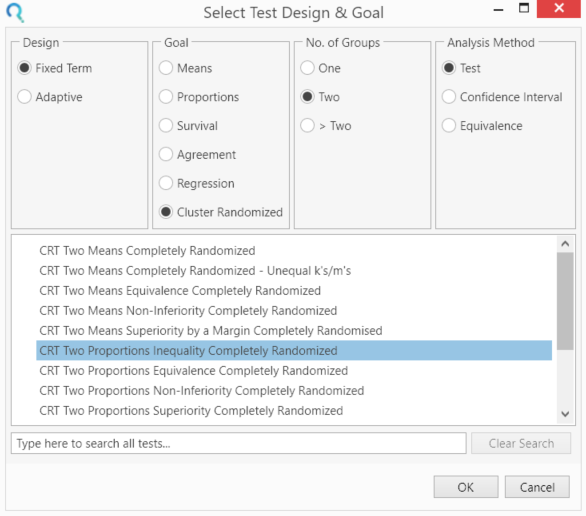
Select the CRT Two Proportions Inequality Completely Randomised table.
This can be done using the radio buttons or alternatively, you can use the search bar at the end of the Select Test Design & Goal window.
Step 2:
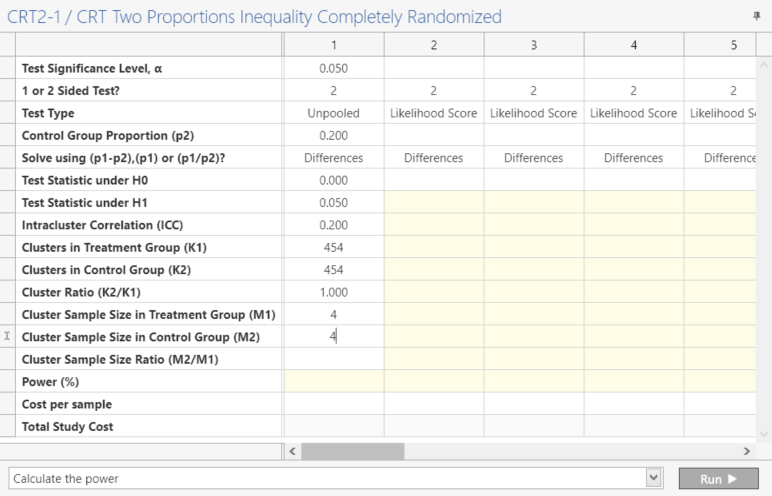
Enter the parameter values for power calculation taken from the study protocol.
Step 3:
Once the parameter values are entered from Step 2, click Run.
| This analysis gives a power of 81.529 / 81.53 as per the targeted power of 80% |
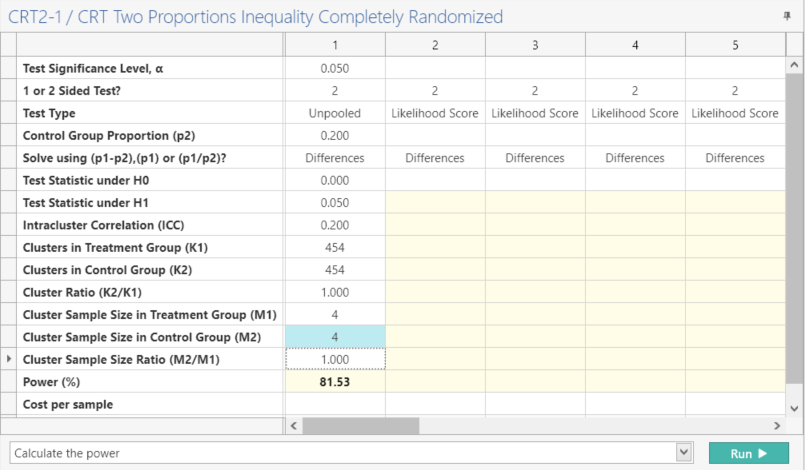
The slight increase in power could be due to rounding or different assumptions
regarding the test statistic used e.g. continuity correction, chi-squared statistic,
sine adjustment.
Note: When using nQuery Advanced both the Cluster Sample Size Ratio and Power (%) will be auto-calculated once all the parameter values from Step 2 are entered.
Step 4:
Once the calculation is completed, nQuery Advanced provides an output statement summarizing the results. It States:
| Output Statement: |
|
“In a cluster randomized trial comparing two binary variables, a sample size of 454 clusters with 4 individuals per cluster in the treatment group and a sample size of 454 clusters per group with 4 individuals per cluster in the control group achieves 81.53% power to detect a difference between two proportions when the Differences under null hypothesis and alternative hypotheses are 0 and 0.05 respectively, the control group proportion is 0.2, the intracluster correlation is 0.2 and when using a two-Sided test at the 0.05 significance level using the Unpooled score statistic.” |

Copyright © Statsols. All Rights Reserved. Do Not Sell or Share My Personal Information. Privacy Policy .